-
Apps and Services
-
LiteMol
A powerful 3D molecular viewer -
PatternQuery
Find atom patterns in molecular databases -
CoordinateServer
Streaming of 3D molecular data -
DensityServer
Streaming of 3D experimental data -
AtomicChargeCalculator II
Compute, compare, and visualize atomic charges -
MotiveValidator
Validate residues, ligands, and molecular fragments -
ValidatorDB
PDB database validated using MotiveValidator -
ValTrendsDB
Discover trends in structure quality -
MOLE 2.5
Compute and analyze channels -
CrocoBLAST
Local sequence alignment
-
- Wiki
- About
PatternQuery
Find atom patterns in molecular databases
PatternQuery is an interactive, user-friendly, and platform independent web service enabling the user to effectively define, extract, and analyze structural patterns or biomolecular complexes using the PatternQuery language. Such analysis is particularly useful not only in the structural and functional assignment of uncharacterized or newly determined proteins, but also represents a key point in rational design and engineering of novel functional sites, and comparative protein structural analyses.
×
What can I do with PatternQuery? See the Samples in the tab below.
PatternQuery is currently available in 3 modes:
- Explorer – a playground for querying small data sets, analyzing patterns, and tweaking queries.
- Service for querying the entire Protein Data Bank.
- Command Line Version for querying custom databases.
The button in the top right corner of the page is a good place to start using the service.
Welcome to PatternQuery submission page. Access different tabs for running the PQ service for the detection of structural patterns over the whole Protein Data Bank or in your own data set of molecular structures.
Different sections of the web page offer interactive guides indicated by
which give a quick walk through all the main elements of the page.
Many tool tips are available by hovering over any graphical or textual element in the interface. If you require further assistance, do not hesitate to contact us via email (david.sehnal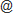 mail.muni.cz)
or use the support tab.
mail.muni.cz)
or use the support tab.
Refer to the Wiki Manual for any clarifications (especially User Interface and the language reference).
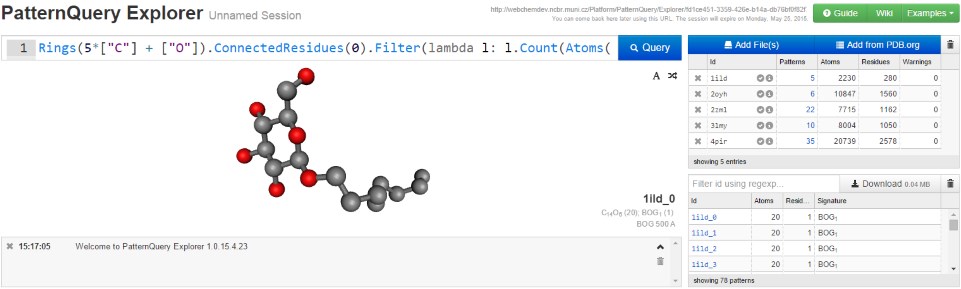
PatternQuery relies on an internal chemical language. The image gives an example of a query composed in the PatternQuery language. It identifies residues containing pyran rings (typically sugars and sugar derivatives), and retrieves each such residue together with all residues within 3Å of it. To learn more about how the language works, you can start with the language principles or this guide describing how to build a query.
If you found PatternQuery helpful, please cite:
Sehnal, D., Pravda, L., Ionescu C.-M., Svobodová Vařeková, R. and Koča, J. (2015)
PatternQuery: web application for fast detection of biomacromolecular structural patterns in the entire Protein Data Bank. Nucleic Acids Res., 43, W383–W388.

PatternQuery is a part of services provided by ELIXIR –
European research infrastructure for biological information.
For other services provided by ELIXIR's Czech Republic Node visit www.elixir-czech.cz/services.
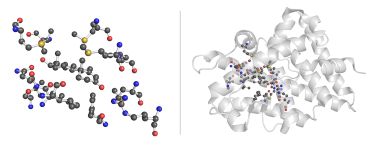
Testosterone Binding Site Browse Data
Testosterone is a steroid sex hormone found in a variety of vertebrates. It activates an androgen receptor (NR3C4) upon binding in either its pure form or its derivative dihydrotestosterone and is primarily responsible for the development of male primary sexual characteristics [1].
Residues("TES") Residues annotated as TES ...
.AmbientResidues(4) ... and all residues within 4Å of the particular TES occurrence.
The Protein Data Bank contains 26 instances of residues annotated as testosterone (TES) originating from 26 PDB entries (as of Dec 23 2014). The testosterone derivatives with different annotation (BDT, DHT, FFA, TH2) were not queried here. All the structures are complete and correct. The immediate surrounding of TES residues is rich in positively charged (Arg, His) and polar residues (Thr, Gln, Tyr).
References:- Voet, D. and Voet, JG. Biochemistry. 4th ed. Hoboken, NJ: John Wiley, 2011, xxv, 142853. ISBN 04-709-1745-8.
Zinc Fingers Cys2His2 Zinc Finger Transcription Factor Browse Data
Cys2His2 zinc fingers (C2H2-ZFs) are the largest family of DNA-binding proteins in metazoans. They provide a stable, versatile and conserved framework for double-stranded DNA recognition [1].
RegularMotifs(".{2}C.{2,4}C.{3}[F|Y].{5}[AILFPGV].{2}H.{3,5}H") Detect specified primary sequence motif ...
.ConnectedAtoms(1) ... and atoms connected to it.
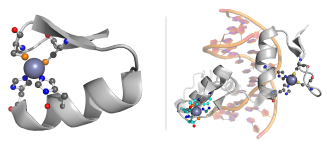
Each zinc finger spans ~30 amino acid residues and folds into a simple ββα-motif around a tetrahedrally coordinated zinc ion. The motif can be identified by a regular expression X2-C-X2,4-C-X12-H-X3,5-H1,2, where X represents any amino acid. The X12 region usually has the form: X3-[F|Y]-X5-Ψ-X1,2, where Ψ denotes a hydrophobic residue [2]. The zinc ion is coordinated by two cysteine residues and two histidines.
PQ was employed in the discovery and extraction of all structural patterns which satisfy the sequence condition. In total, 354 patterns representing zinc fingers were successfully identified in 233 distinct PDB entries. The majority of patterns come from Homo sapiens and other mammals.
References:- Gupta, A., Christensen, R.G., Bell, H.A., Goodwin, M., Patel, R.Y., Pandey, M., Enuameh, M.S., Rayla, A.L., Zhu, C., Thibodeau-Beganny, S., et al. (2014) An improved predictive recognition model for Cys(2)-His(2) zinc finger proteins. Nucleic Acids Res., 42, 4800–12.
- Pabo, C.O., Peisach, E. and Grant, R.A. (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem., 70, 313–40.
LecB Sugar Binding Sites Browse Data
Pseudomonas aeruginosa is an opportunistic pathogen associated with a number of chronic infections. This pathogen forms a biofilm shield enabling it to survive both the response of the host immune system and antibiotic treatment. One of the cornerstones of biofilm formation is the presence of carbohydrate-binding proteins (lectins) on the outer cell membrane: LecA (PA-IL) and LecB (PA-IIL). Their inhibition is considered to be a promising approach for anti-pseudomonadal treatment [1].
Near(4, Atoms("Ca"), Atoms("Ca")) Pairs of Ca atoms closer than 4Å ...
.ConnectedResidues(1) ... with residues connected to any one of them ...
.Filter(lambda l: ... the residues must contains a sugar ring (pyran, furan) ...
l.Count(Or(Rings(5 * ["C"] + ["O"]), Rings(4 * ["C"] + ["O"]))) > 0)
.Filter(lambda l: l.Count(Atoms("P")) == 0) ... and no P atoms (to exclude nucleotides).
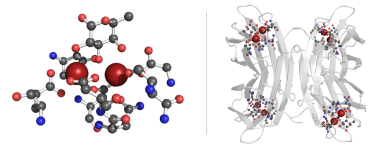
We employed MQ in the discovery of sugar binding sites with similar geometry as the tetrameric PA-IIL protein in the Protein Data Bank. The carbohydrate-binding domain is calcium dependent, with two calcium ions stabilizing the binding site and contributing to sugar binding [2]. Therefore, just the structures containing calcium ions were queried. The query was searching for 2 calcium ions at most 4Å far from each other, and all the residues with direct interaction with either of the ions. Furthermore, just the molecular patterns containing a furan or pyran ring were kept. Additionally, in case the phosphorus atom was present, the pattern was filtered out in order to avoid nucleotides.
The outcome of this query is made up of 87 distinct patterns originating from 36 PDB entries of 7 different organisms. The majority of them originated from P.aeruginosa, however, binding sites from other pathogens such as R. solanacearum, B. cenocepacia or C. violaceum were identified. In 83 patterns, the sugar-binding site is composed of 3x Asp, 2x Asn, 1x Glu and 1x Gly residues, in agreement with literature reports of the binding site for 24 PDB entries belonging to 3 pathogens.
References:- Hauck, D., Joachim, I., Frommeyer, B., Varrot, A., Philipp, B., Möller, H.M., Imberty, A., Exner, T.E. and Titz, A: Discovery of two classes of potent glycomimetic inhibitors of Pseudomonas aeruginosa LecB with distinct binding modes. 2012, ACS Chem. Biol., 8, 1775–84.
- Mitchell, E., Houles, C., Sudakevitz, D., Wimmerova, M., Gautier, C., Pérez, S., Wu, A.M., Gilboa-Garber, N. and Imberty, A: Structural basis for oligosaccharide-mediated adhesion of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. 2012, Nat. Struct. Biol., 9, 918–21.
PatternQuery Service
Use the PatternQuery Service to search for structural patterns throughout the entire Protein Data Bank.
×
To begin, use the
and see the examples
and the language reference. If you need more help, feel free to
ask.
The PDBe.org database mirror was last updated on
and contains entries
(with metadata).
Obsolete entries are not included.
Our PDBe.org database mirror does not contain .
These entries either do not exist, are obsolete, or were added very recently.
A list of PDB entry identifiers, separated by a new line or a comma
(e.g. 1tqn,3d12).
For example, you may paste a list of pre-filtered PDB IDs (e.g., by organism, molecular weight, etc,) from PDB.org:
- Go to PDB.org Search and enter your search criteria.
- Run the search and list the results using the option 'Reports: List selected IDs'.
- Copy-paste the result into the text area above.
Current filters match 0 entries in the database. Please update the filter set.
| Category | Comparison | Value |
|---|
Validate ligands and non-standard residues with more than 6 atoms for structure integrity (missing rings/atoms) and stereochemistry.
Encountered some problems:
You are being redirected to the result. This process can take a few seconds depending on the speed of the connection.
If it's taking too long, click here.
PatternQuery Explorer
Use PatternQuery Explorer to search for structural patterns in small datasets or snippets of the PDB, to analyze the patterns, or just to tweak queries that you plan to run on the entire PDB.
PatternQuery Support
If you need help constructing a query or just have a general question, feel free to use this form. We will try to get back to you as soon as possible,
definitely sooner than never.
Your request for support will be assigned a unique URL.
If you provide an email address, we will be able to notify you when we respond to your request. Otherwise, you will just have to come back to that URL and check for an answer. Your email won't be shared with any 3rd party.
Your request for support will be assigned a unique URL.
If you provide an email address, we will be able to notify you when we respond to your request. Otherwise, you will just have to come back to that URL and check for an answer. Your email won't be shared with any 3rd party.
PatternQuery Command Line Version
Run the PatternQuery Service on custom databases. For usage instructions, please consult the
Wiki page.
Current Version (1.1.25.8.19)
Example Data & Configuration
All Versions (change log):
Download
PyMOL Visualization Plug-in
A simple PyMOL plug-in for generating images similar to the ones shown in the Samples tab.
Download the Plug-in